MammOnc-DB is web-platform designed for in-depth analysis of various breast cancer RNA-sequencing and proteomic studies. This platform enables users to access, analyze, and visualize gene expression (RNA-sequencing), epigenetic (ChIP-sequencing), and proteomics (MS-based approach) data from public repositories such as TCGA, NCBI GEO, ProteomeXchange, and PRIDE. Focused on treatment and metastasis in breast cancer, MammOnc-DB offers the following functionalities:
A)
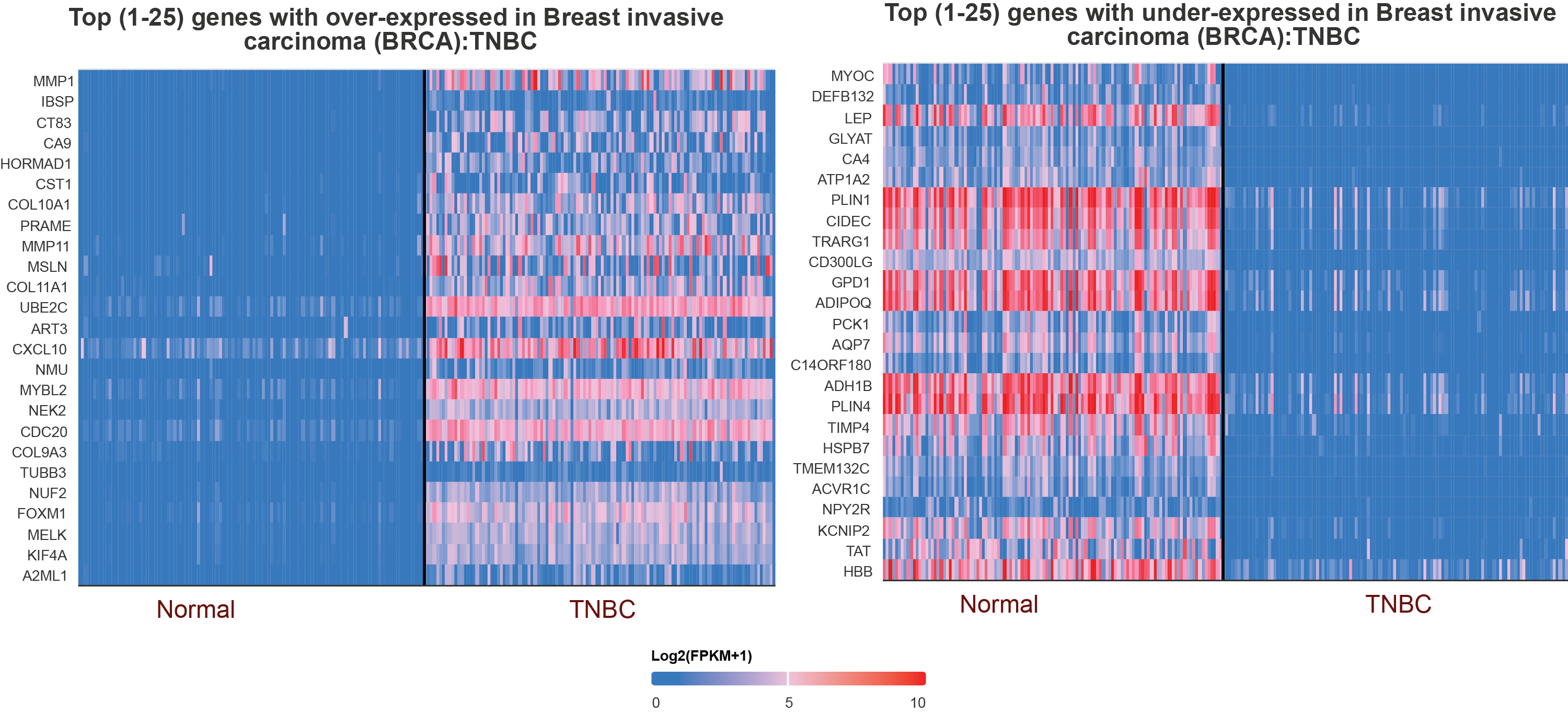
Gene Expression Analysis: Perform relative expression analysis of query genes (mRNA, miRNA, and lncRNA) or proteins as well as single cells sequencing analysis
B)
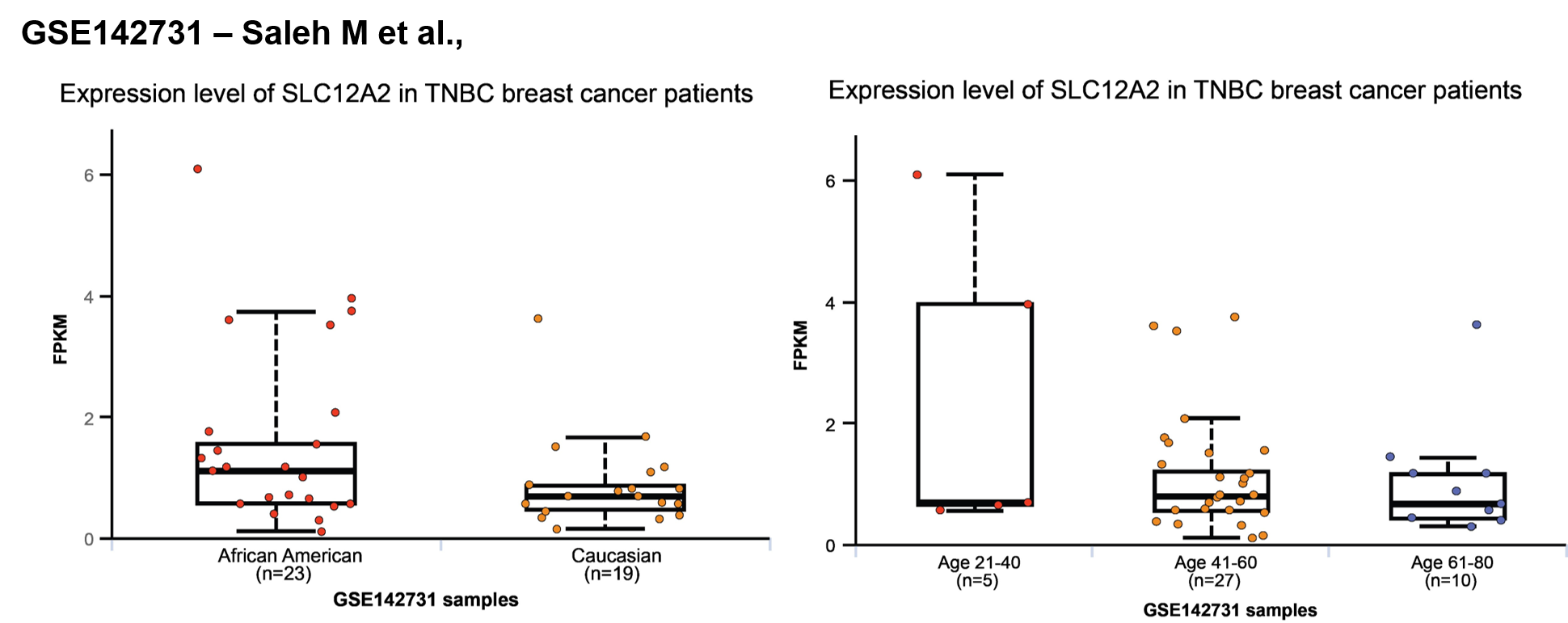
Clinicopathologic Correlation: Determine the impact of gene or protein expression on various clinicopathologic features,
C)
Survival Analysis: Assess the effect of gene expression on survival rates,
D)
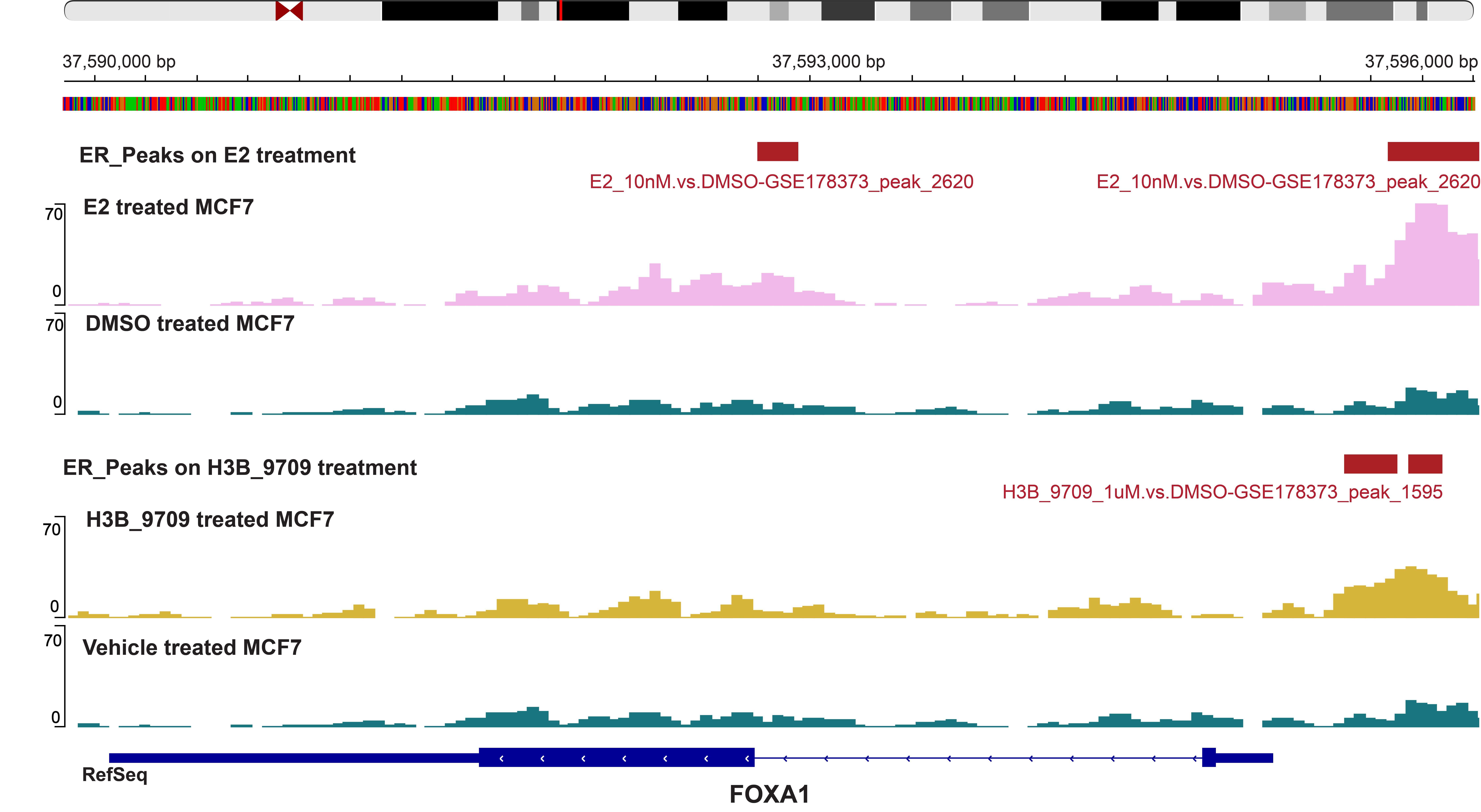
Epigenetic data analysis: Visualize epigenetic changes associated with breast cancer,
E)
Comprehensive Information: Provide additional information about selected genes/targets. The findings are displayed using diverse plots and are available for download in high-resolution formats.
MammOnc-DB aim is to create a global user base that accelerates the discovery of biomarkers in breast cancer. This platform enables biomarker and therapeutic target identification.
Normalization Methods:
Please note that variations in data normalization methods may affect the comparability and interpretation of results. We recommend users consider these differences when analyzing datasets processed through diverse normalization techniques.This work is supported by Breast Cancer Research Foundation of Alabama

Supported in part by NIH/NCI - Comprehensive Partnerships to Advance Cancer Health - 5U54CA118948
Please cite:
Karthikeyan SK, Chandrashekar DS, Sahai S, Shrestha S, Aneja R, Singh R, Kleer CG, Kumar S, Qin ZS, Nakshatri H, Manne U, Creighton CJ, Varambally S.
MammOnc-DB
, an integrative breast cancer data analysis platform for target discovery.NPJ Breast Cancer.
2025 Apr 18;11(1):35. doi: 10.1038/s41523-025-00750-x.[PMID: 40251157]Pan-cancer data analysis platform is at UALCAN (https://ualcan.path.uab.edu)