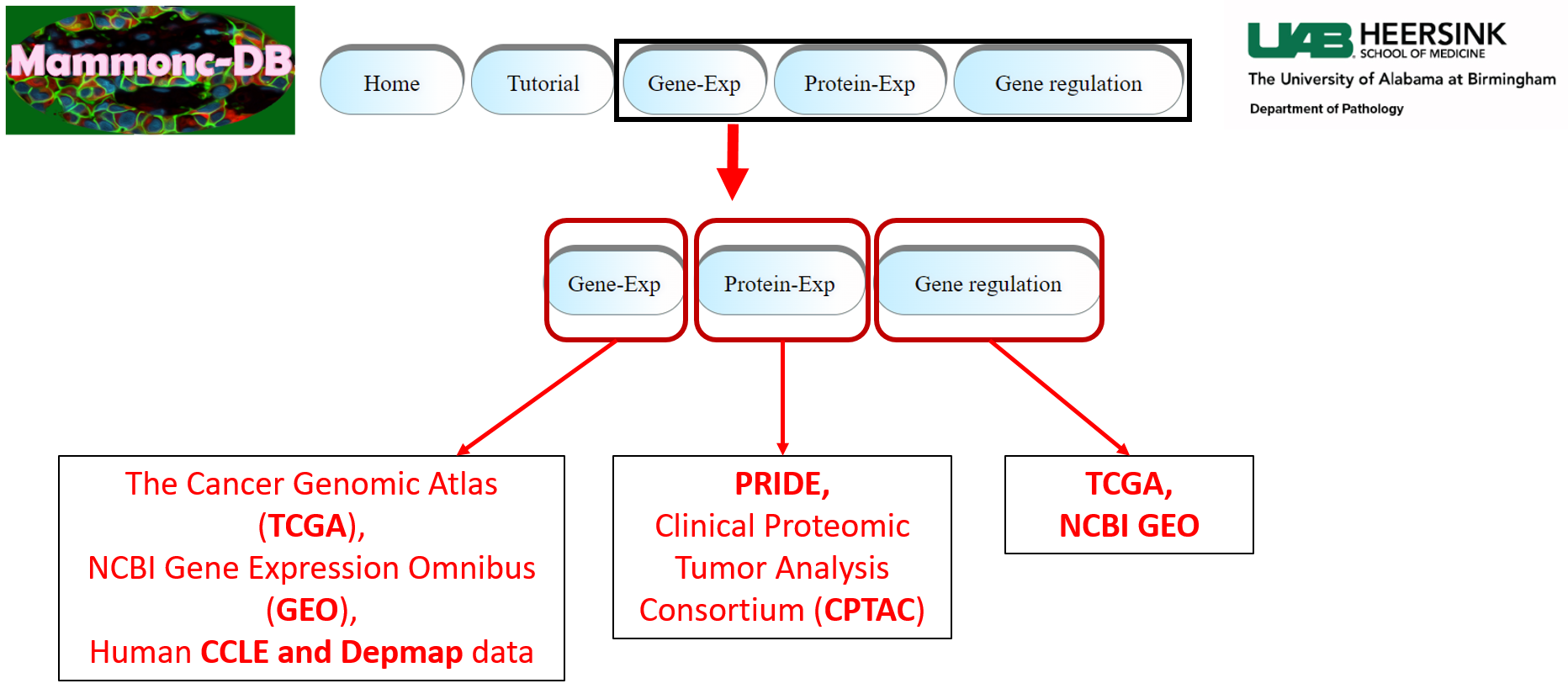
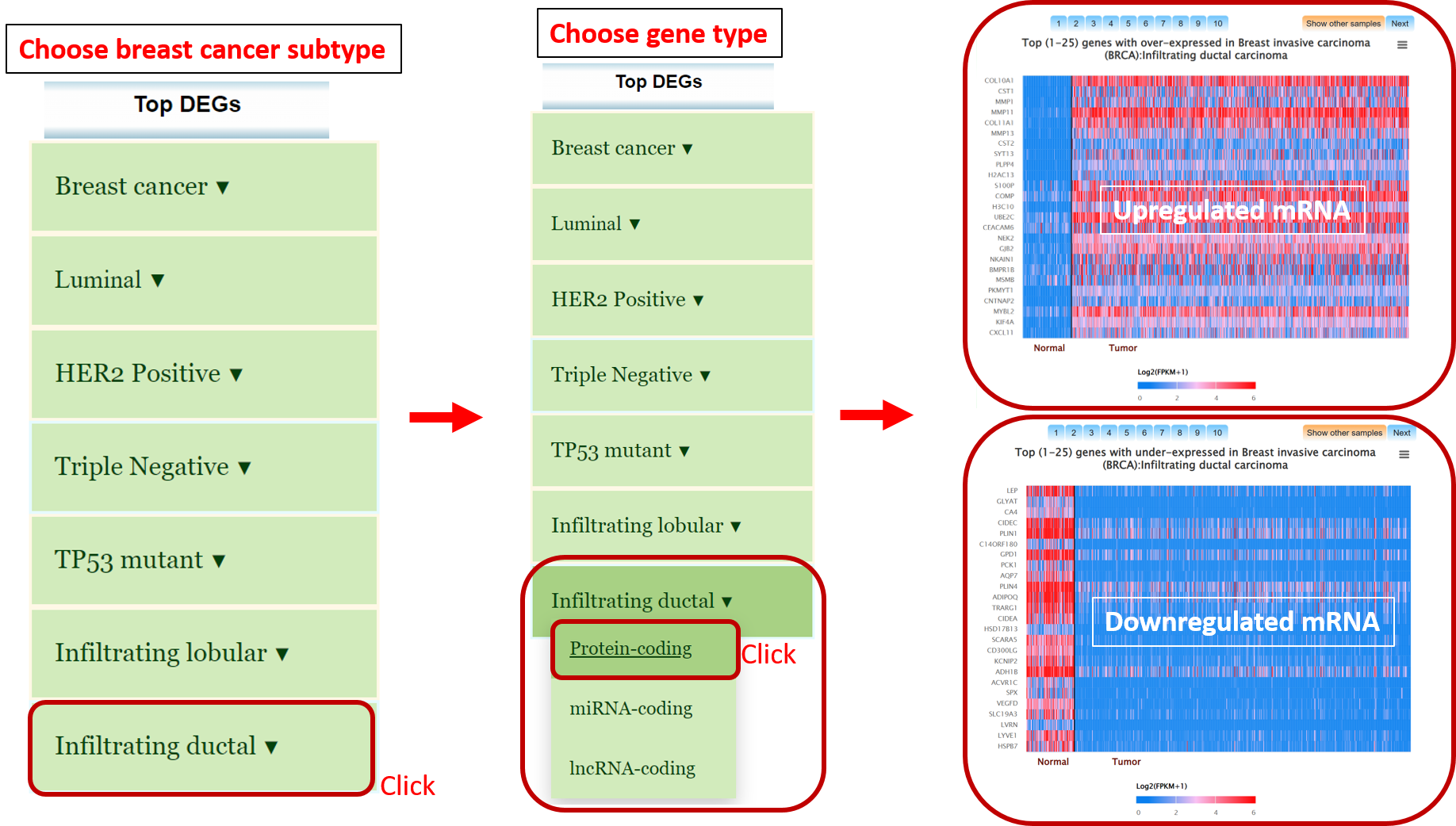
Step 1: Go to menu bar and chose specific omics and chose specific study of interest.
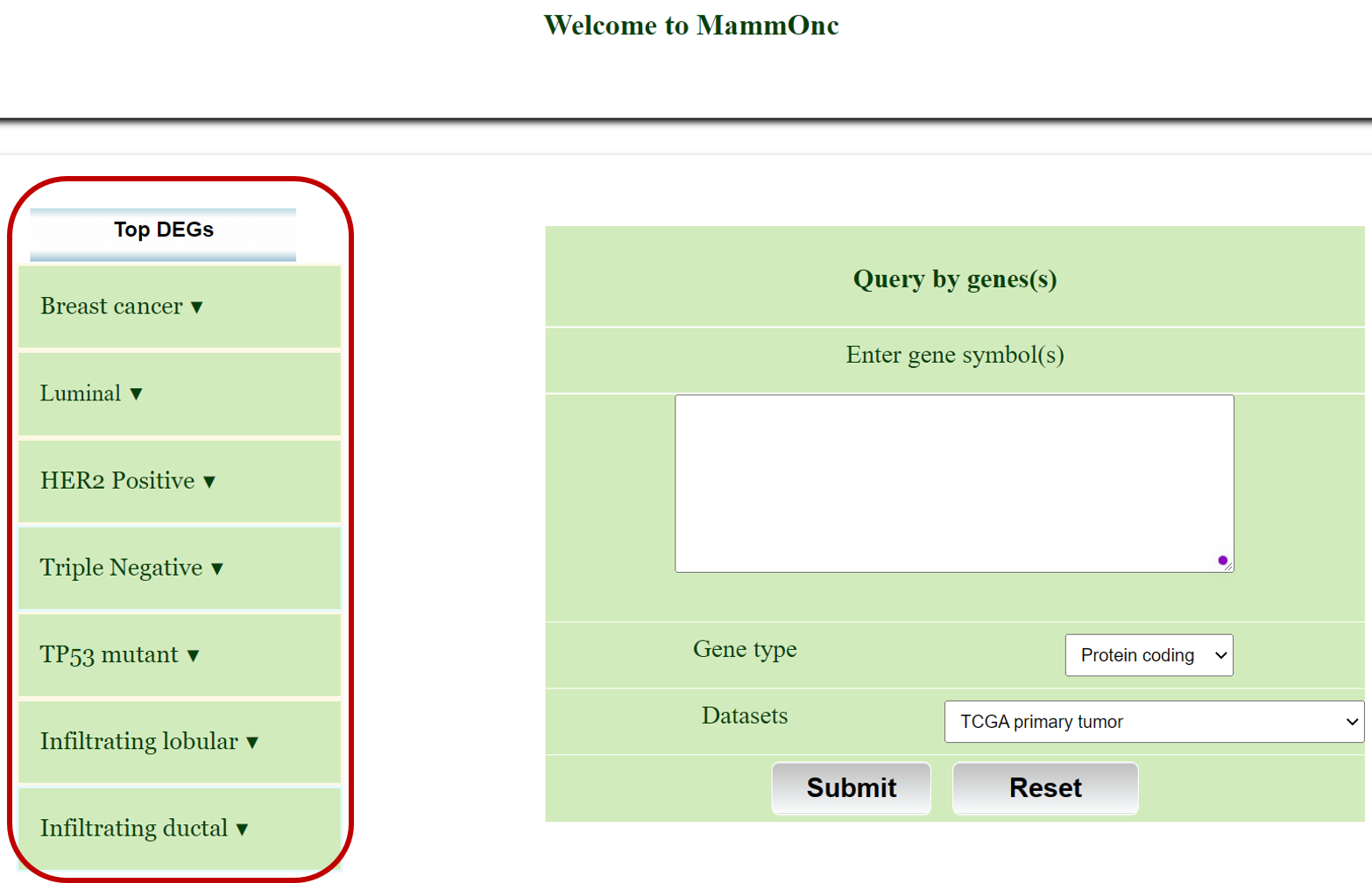
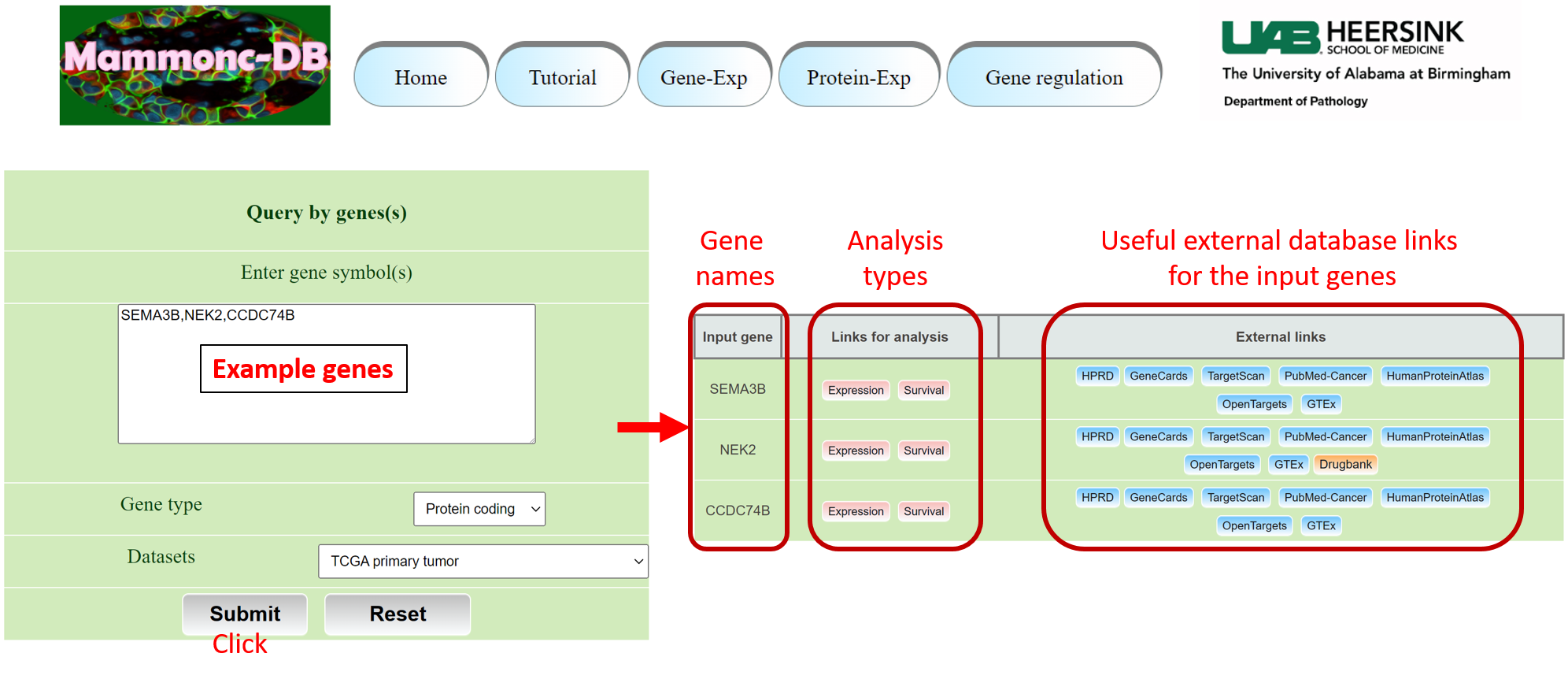
Go to gene expression page of Mammonc-DB and consider left panel for analysis.
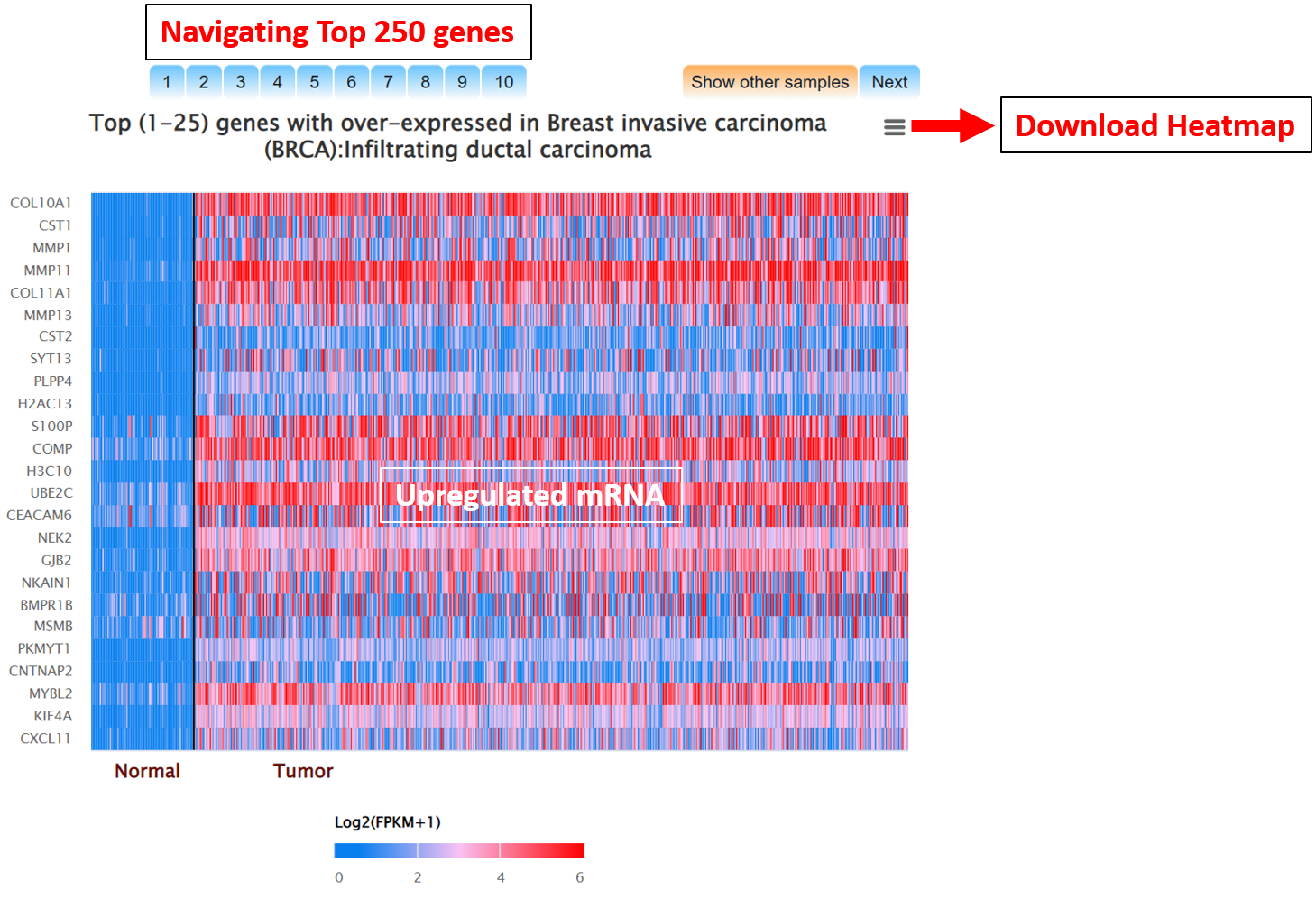
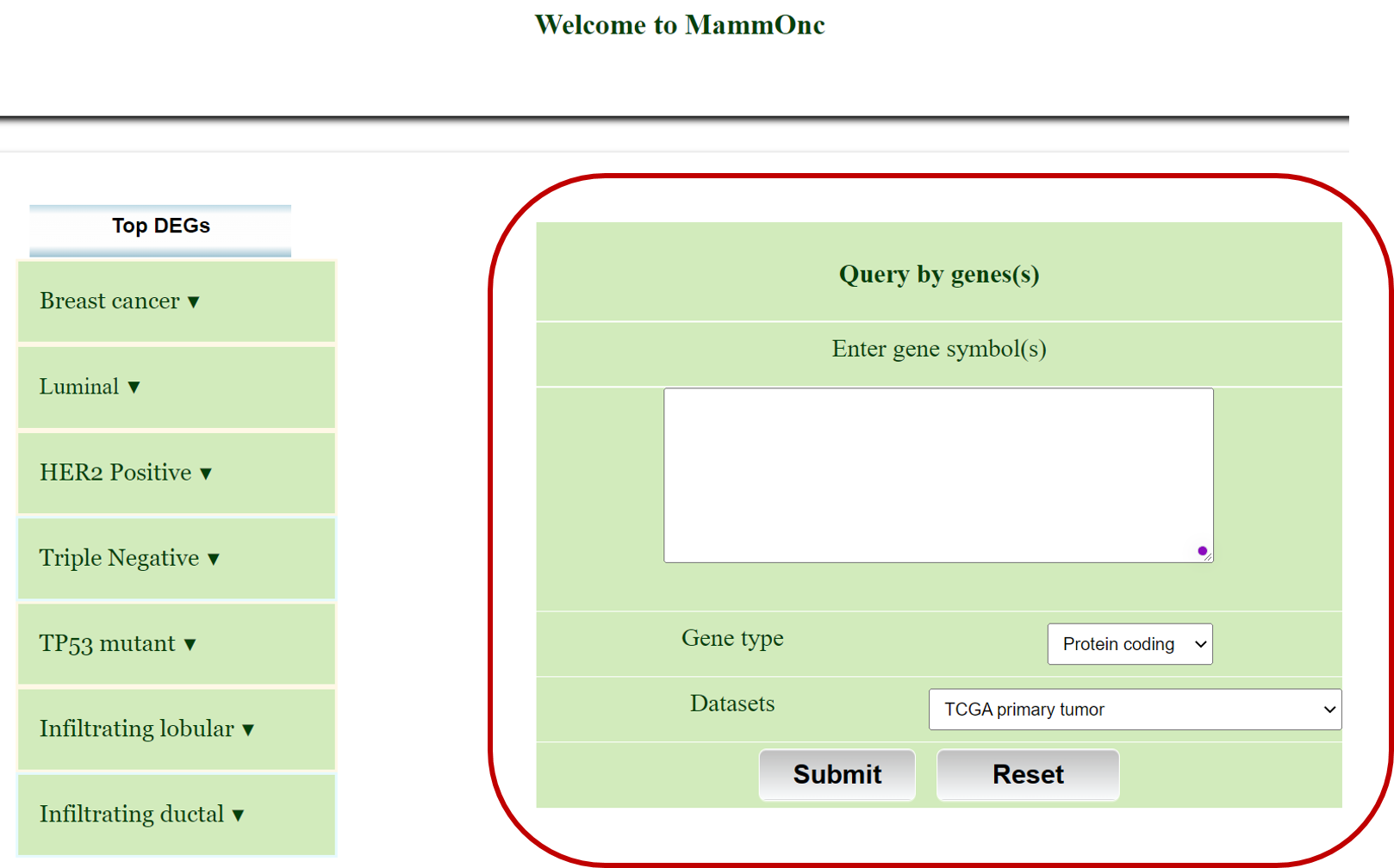
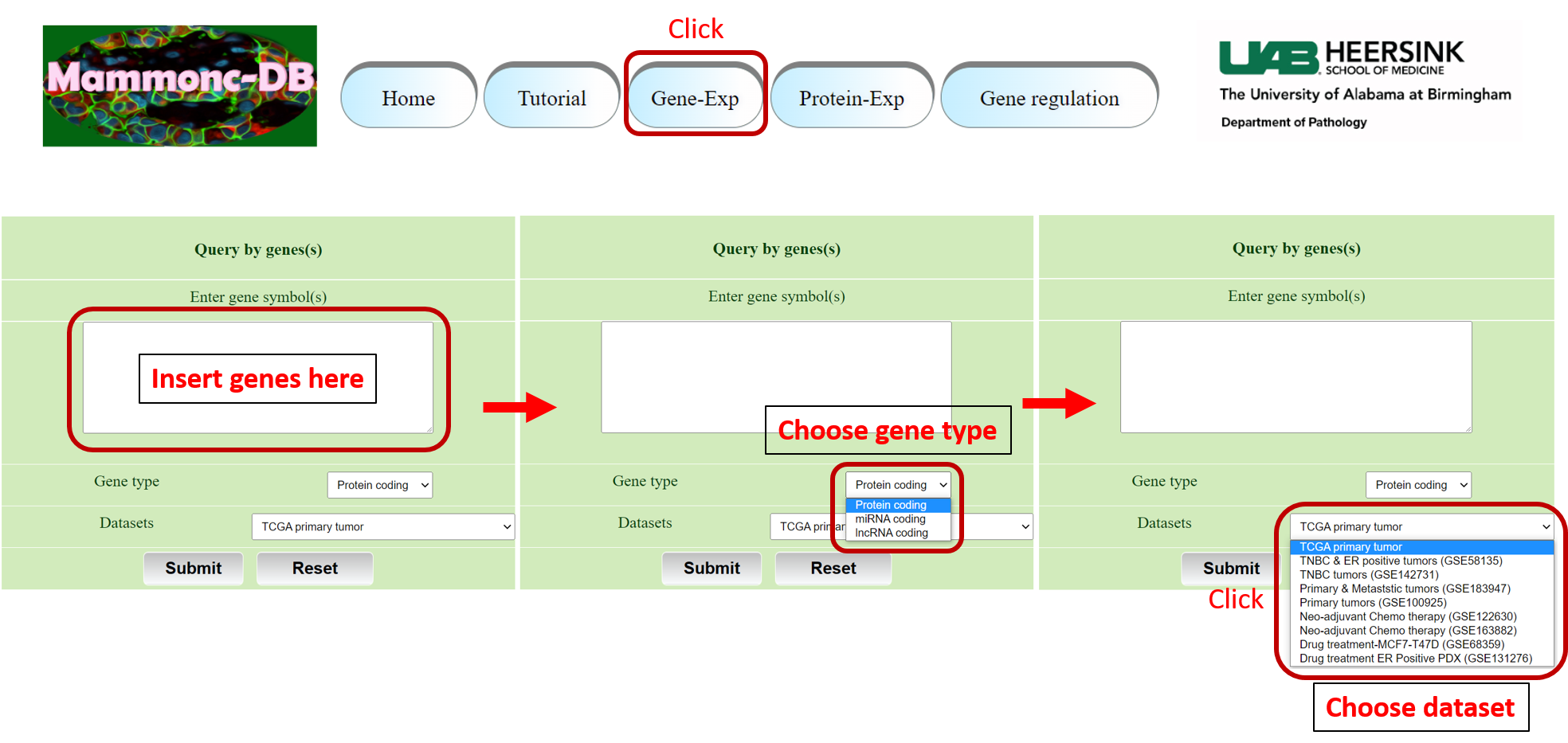
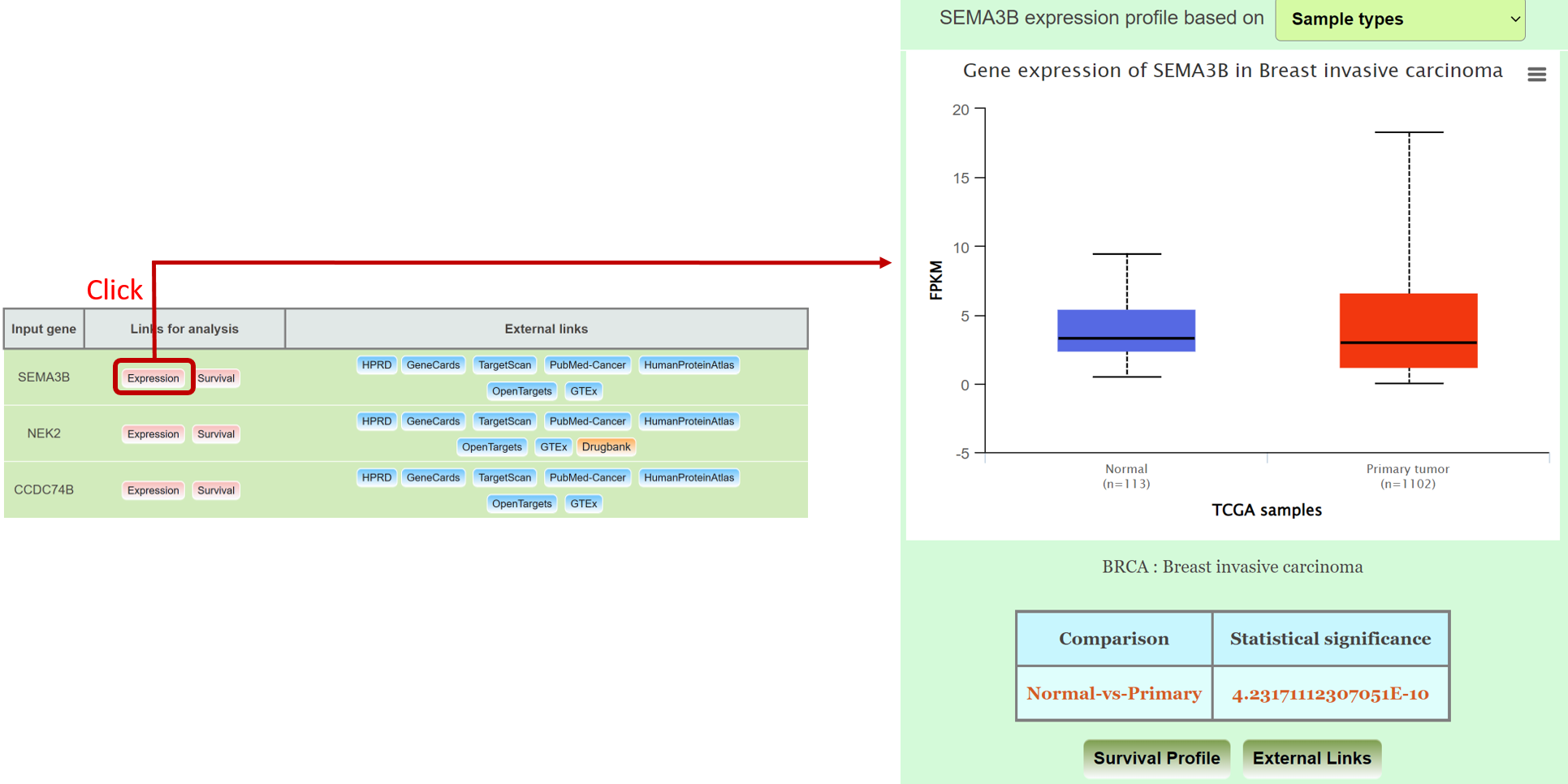
Step 3: Go to gene expression page of Mammonc-DB and consider right panel for analysis.
Example
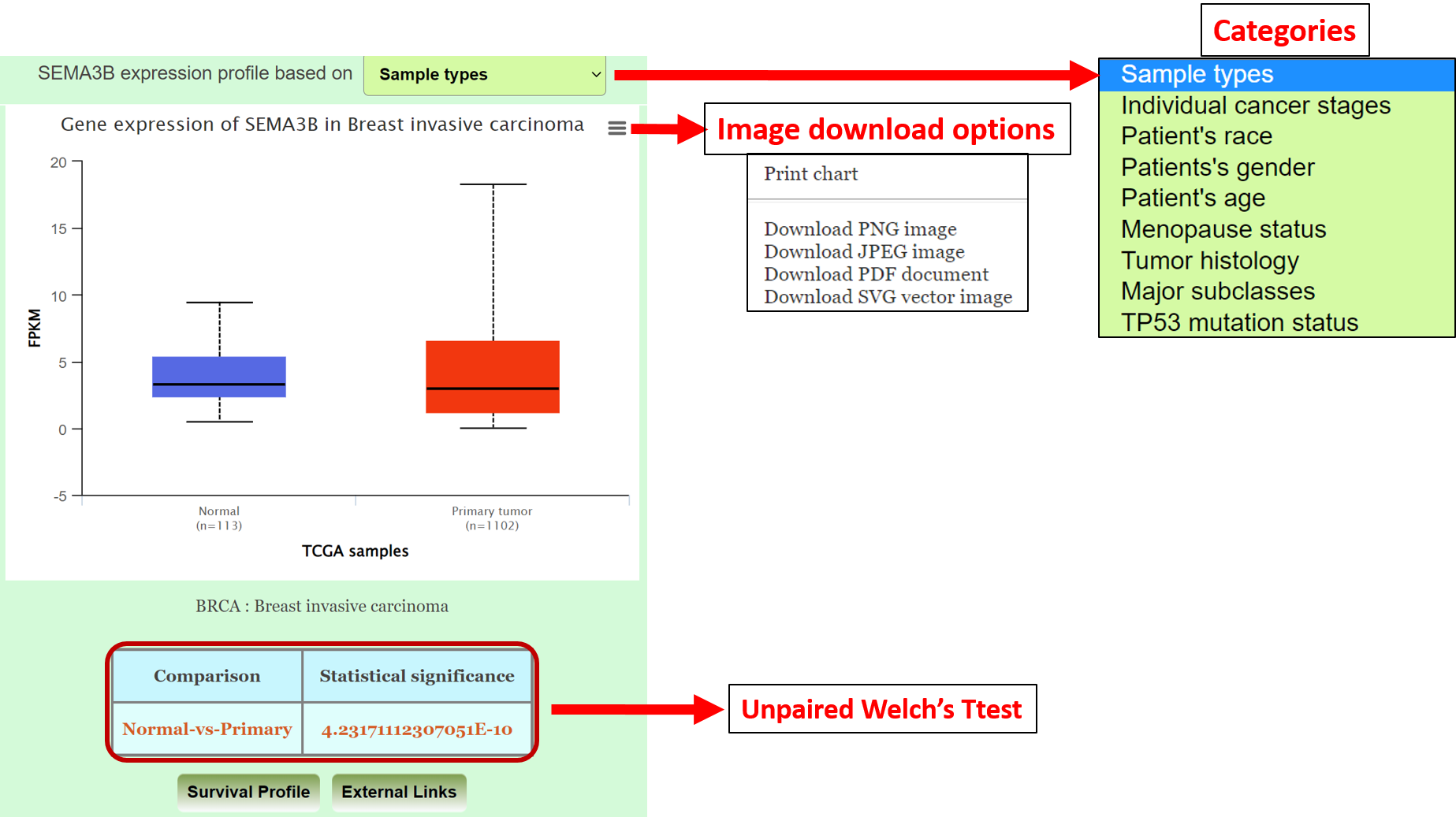
3a: How to explore expression profile of gene of interest based on clinico-pathologic factors?
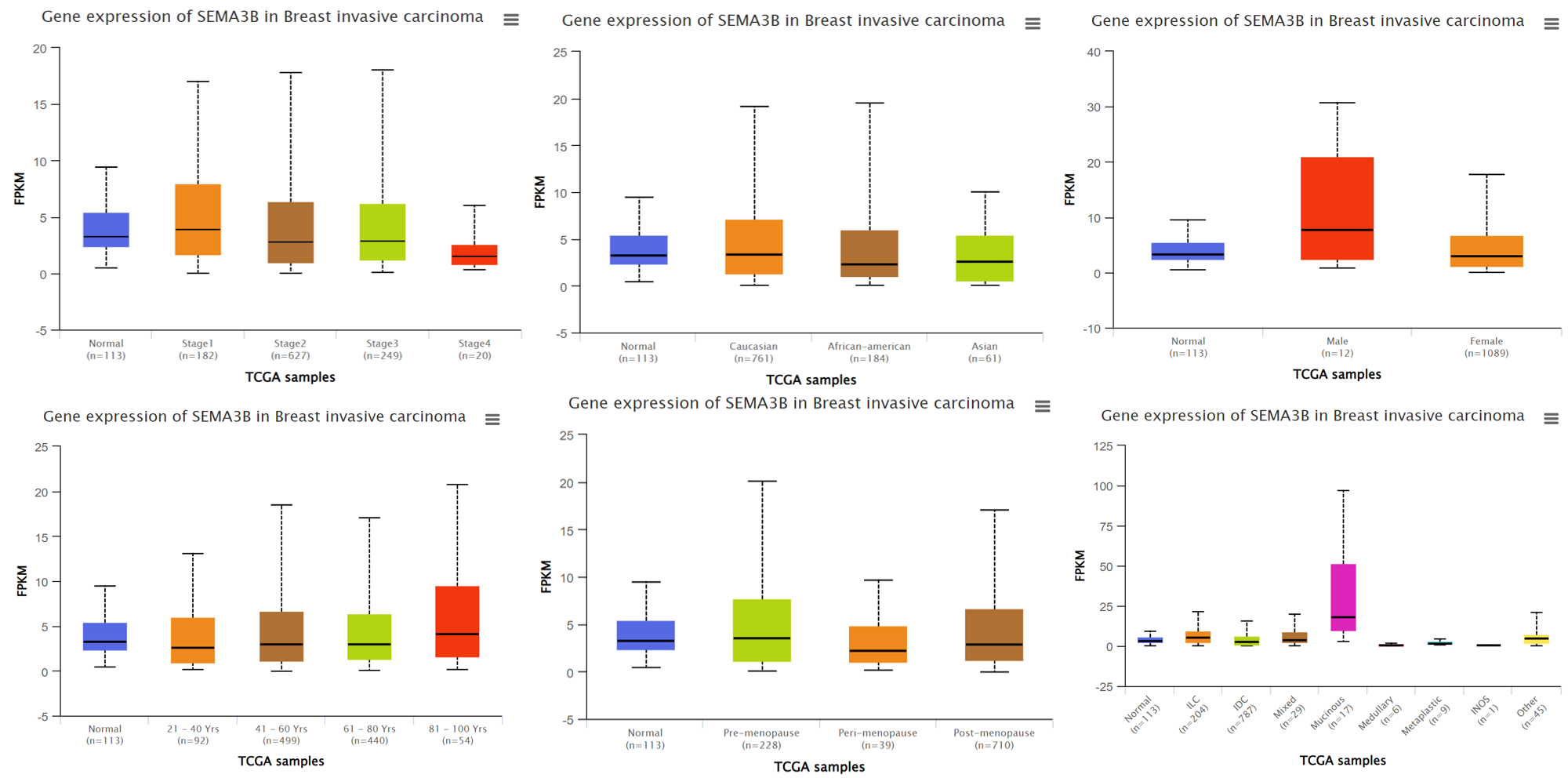
Example of SEMA3B expression based on different clincial features in TCGA.
3b: How to obtain Kaplan meier plot from Mammonc-DB?
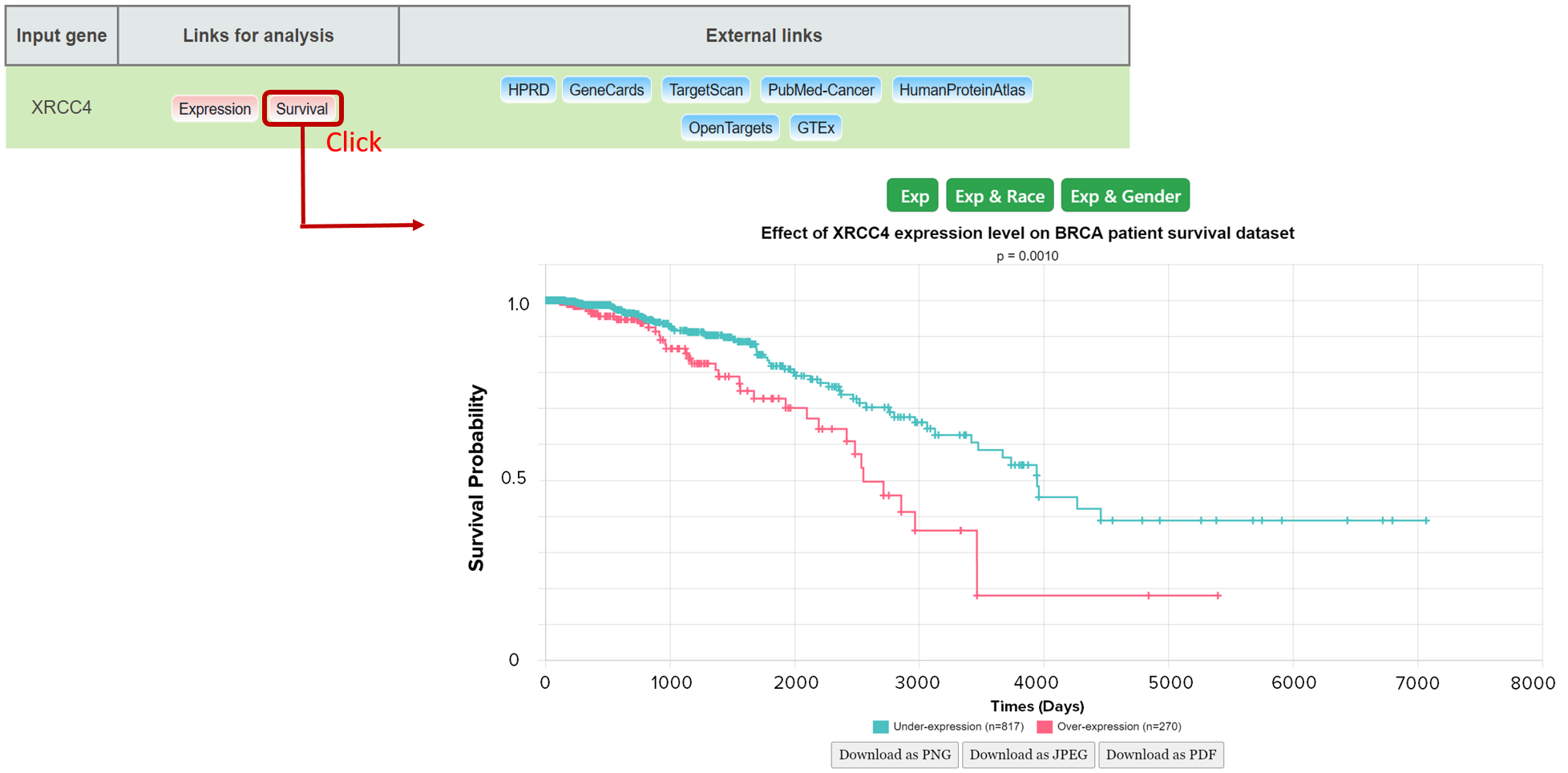
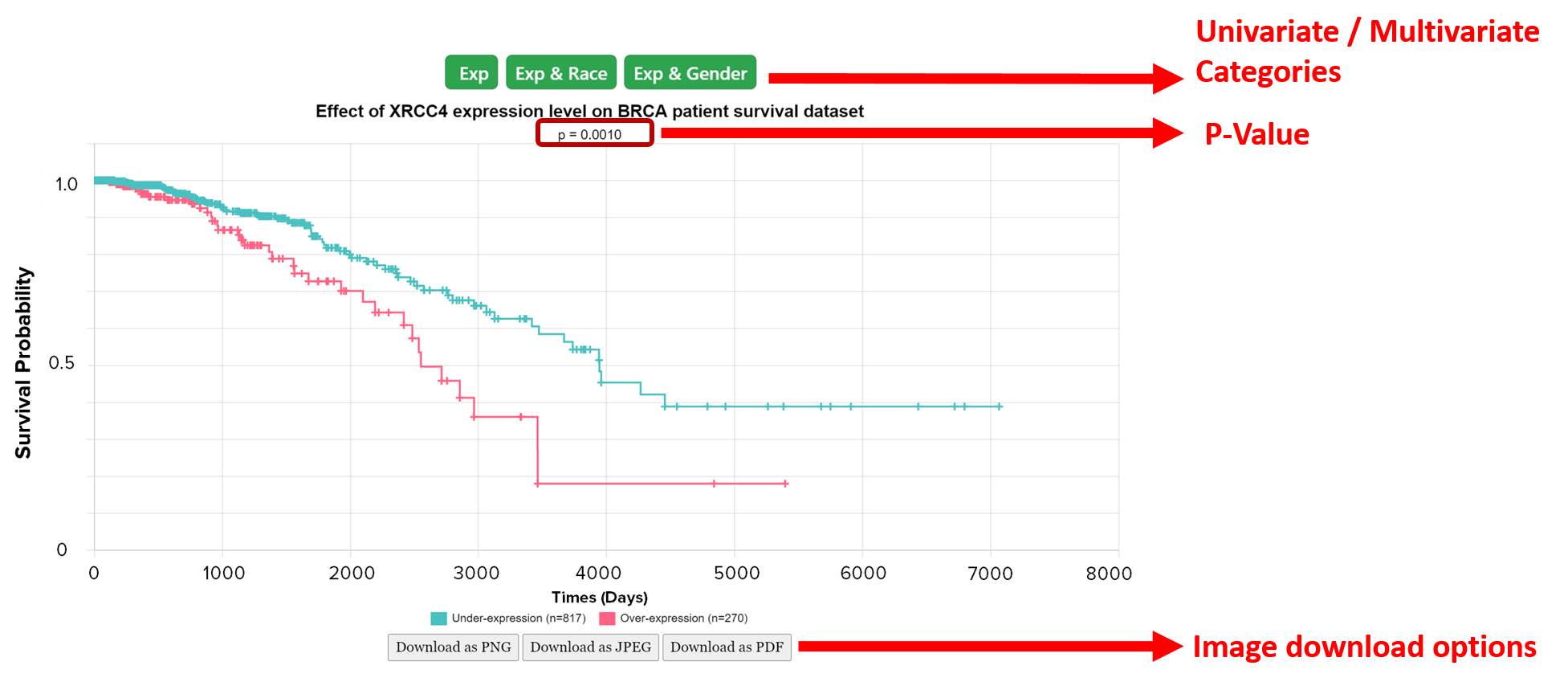
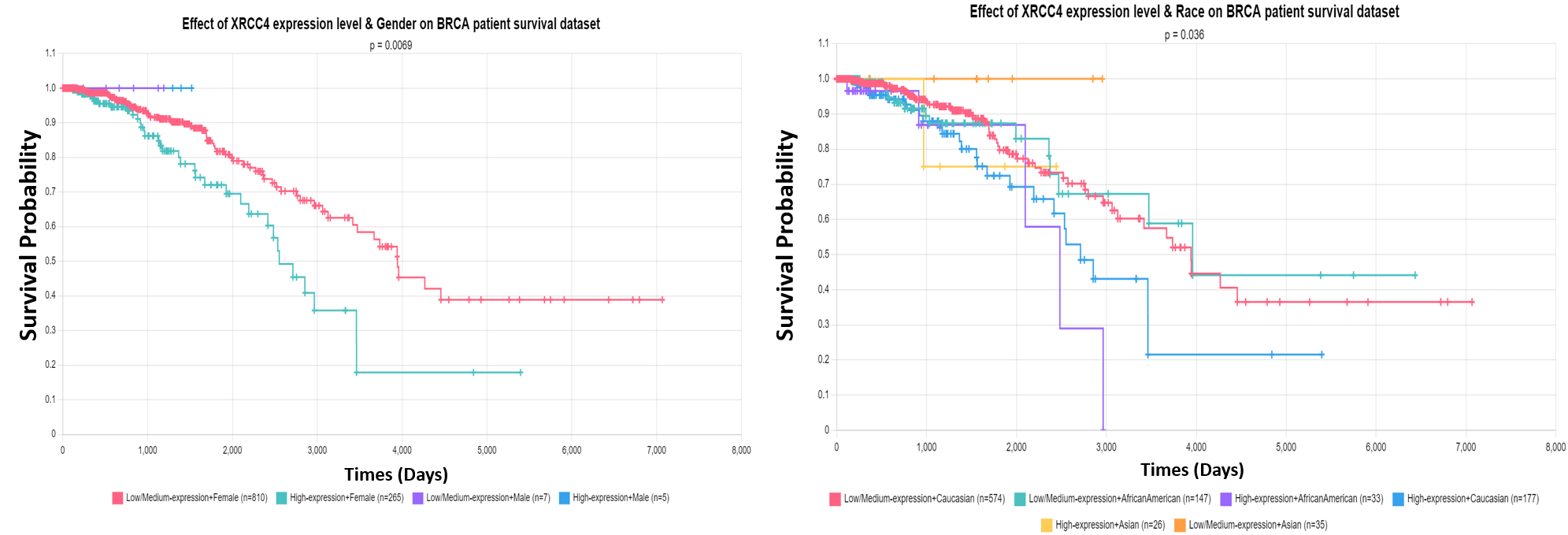
Example of XRCC4 survival analysis based on different clincial features available in TCGA.
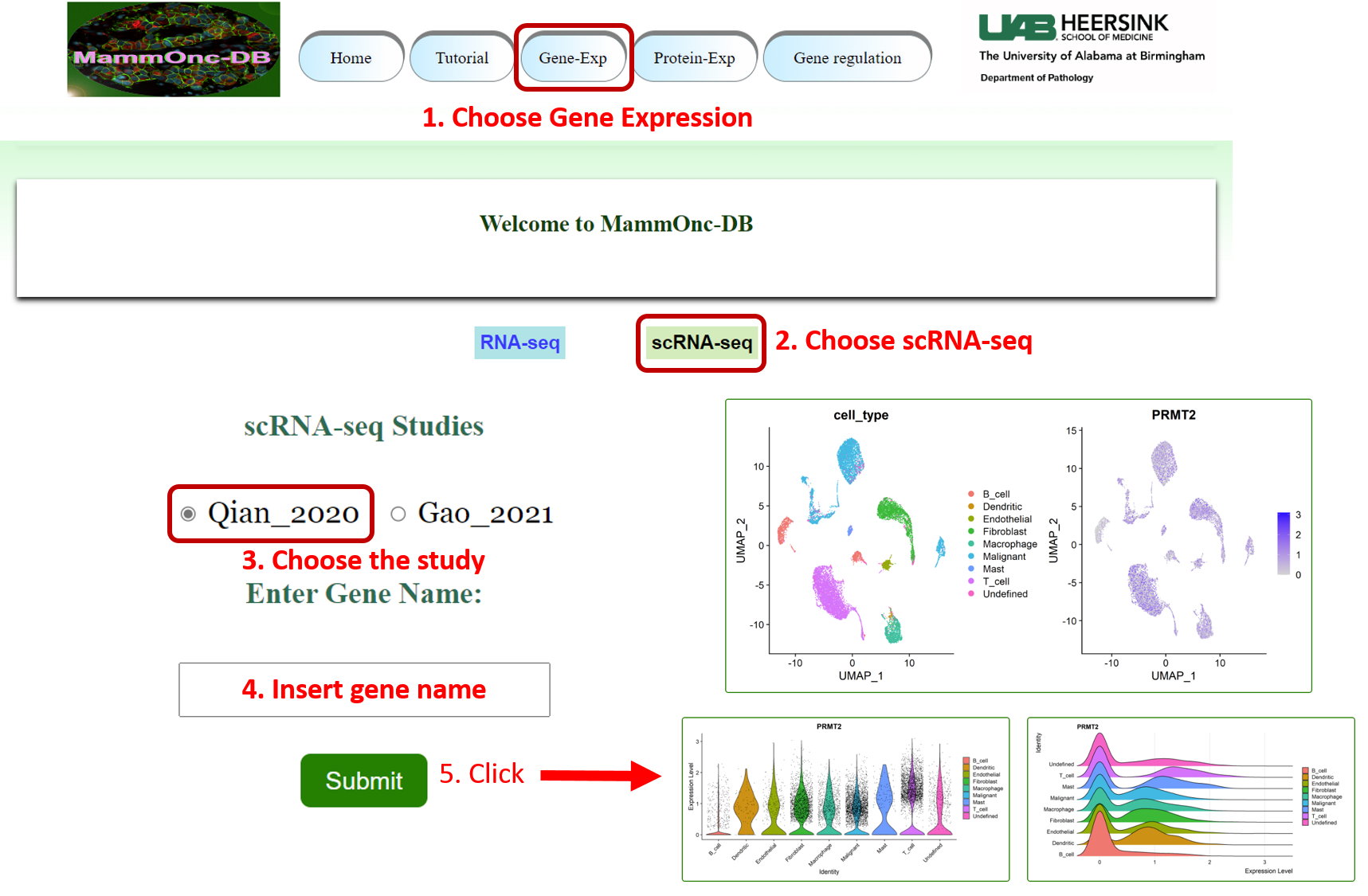
Step 4: Go to gene expression page of Mammonc-DB and consider scRNA-seq button on top for analysis.
Step 5: Go to gene regulation page of Mammonc-DB and consider the panel for analysis.
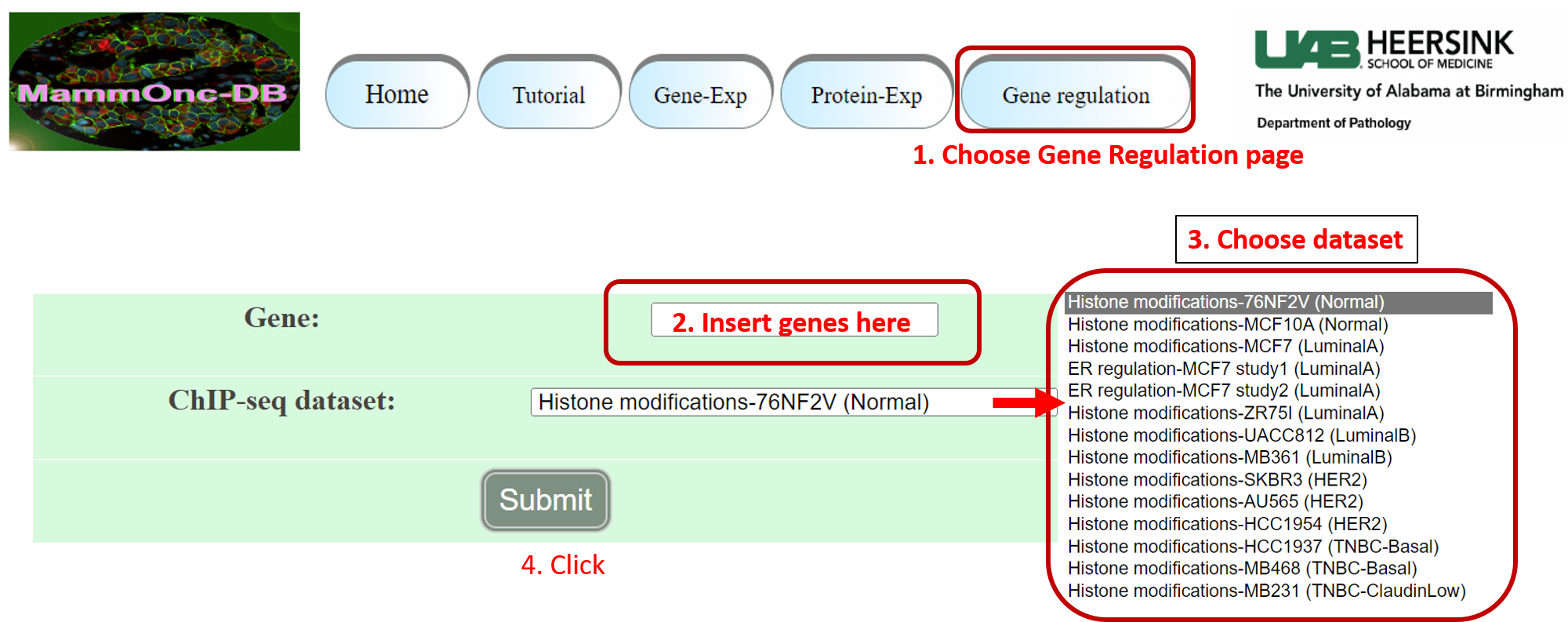
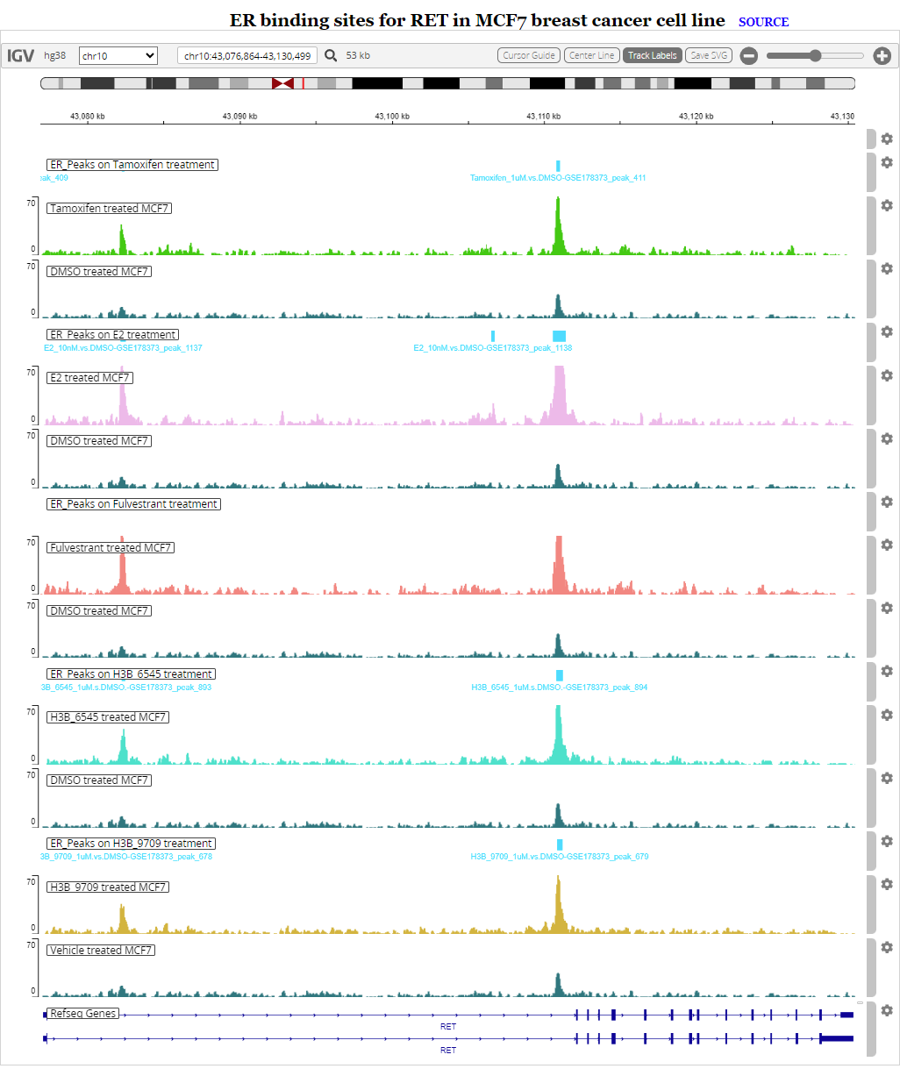
Example of ER binding sites for RET in MCF7 breast cancer cell line.
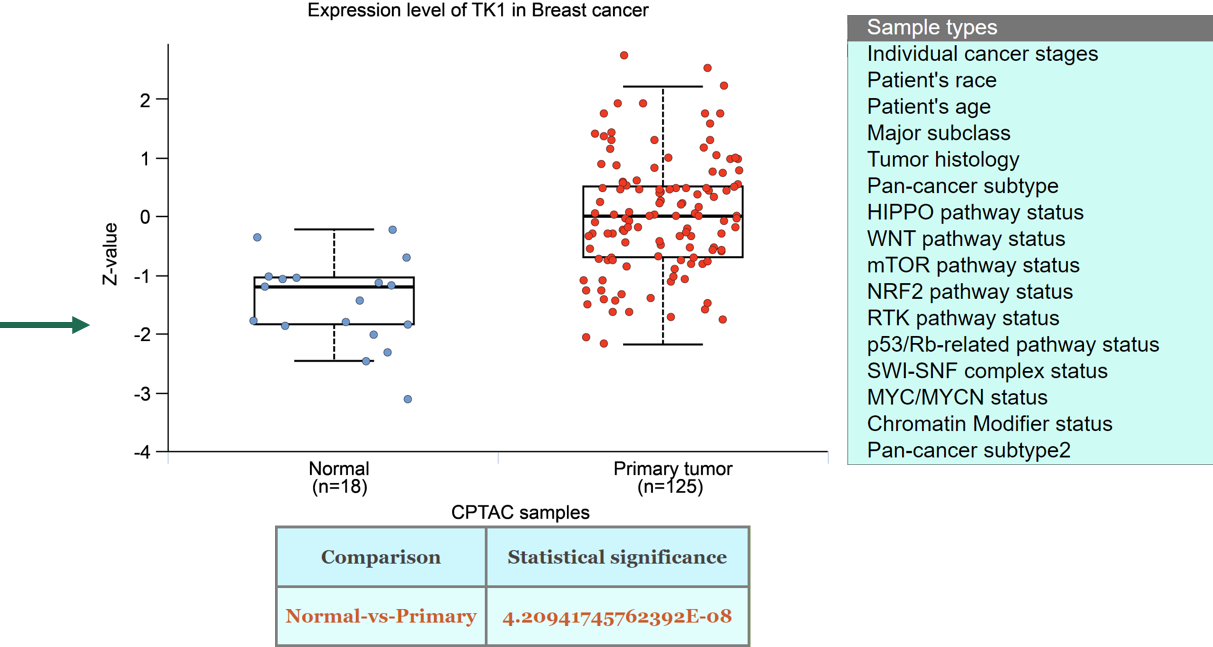
Step 6: Go to protein expression page of Mammonc-DB and consider the panel for analysis.
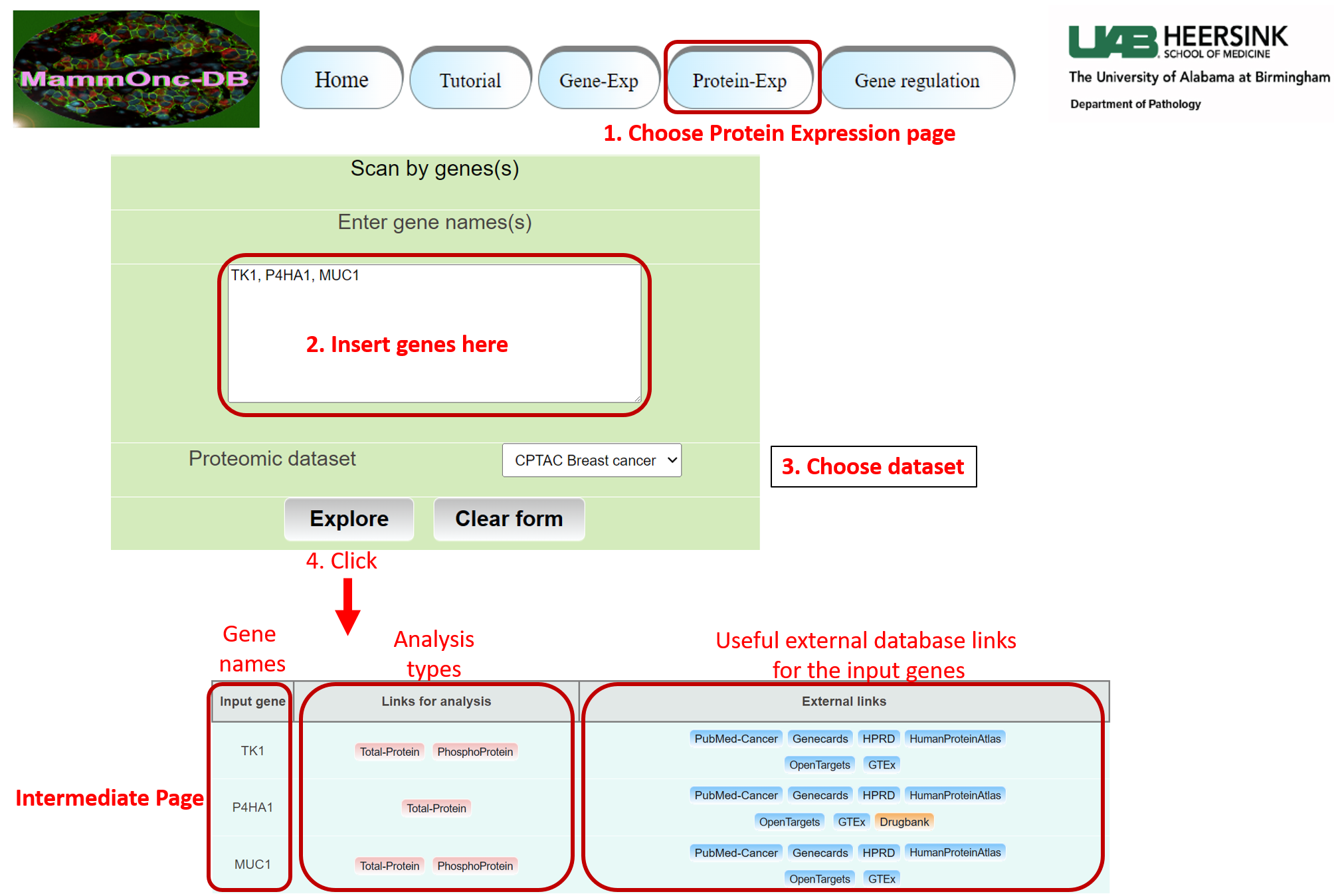
Example of TK1 expression and classifications associated.